Deep learning in population genetics
A review of the state-of-the-art
Population genetics is quickly transitioning into a data-driven discipline. This revolution is happening thanks to the availability of high-throughput genomic data coupled with the need to study increasingly complex evolutionary scenarios.
With likelihood and Bayesian approaches becoming either intractable or computationally unfeasible, machine learning, and in particular deep learning, algorithms are emerging as suitable techniques for population genetic inferences. Deep learning algorithms learn non-linear relationships between the input data and the model parameters being estimated through representation learning from training data sets.
In this review, together with Kevin Korfmann and Oscar Gaggiotti, we illustrate all deep learning algorithms currently employed in population genetics. The paper is available for download here. Specifically, we describe discriminative and generative models with fully connected, convolutional, or recurrent layers. Additionally, as training is performed through synthetic data sets, we list all available simulators to generate training data under complex scenarios. We show how the application of deep learning to empirical data sets mostly replicates previous findings of demography reconstruction and signatures of natural selection in model organisms.
To showcase the feasibility of deep learning to tackle new challenges, we designed a branched architecture to detect signals of recent balancing selection from temporal haplotypic data. Via simulations, we show how the new architecture has good predictive power and how the addition of temporal data enhances the power to detect balancing selection.
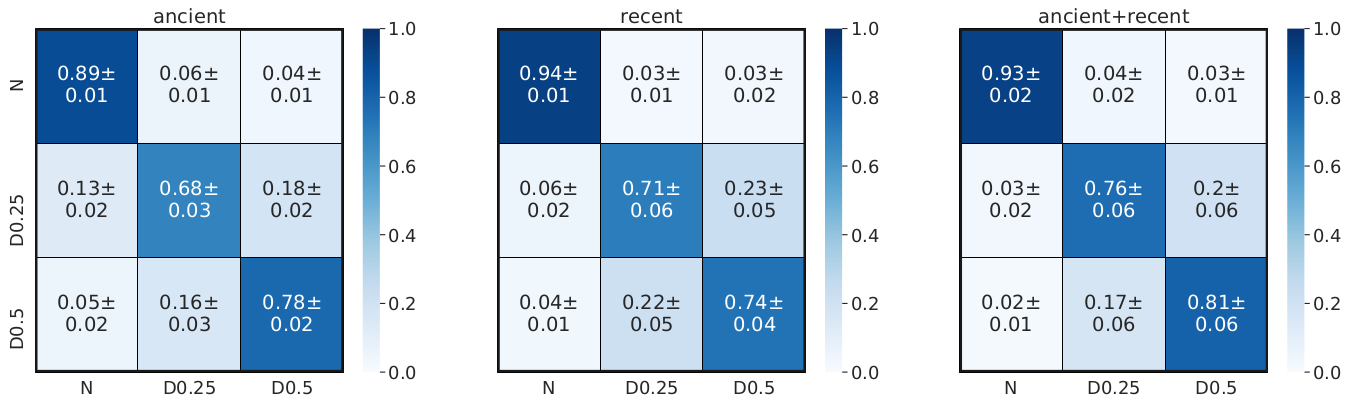
Investigations on the interpretability of neural networks, their robustness to uncertain training data, and creative representation of population genetic data, will provide further opportunities for technological advancements in the field.
We finally discuss how more research is needed in the domain of interpretable machine learning to gain an understanding of how deep learning algorithms make their decisions. This knowledge would enable population geneticists to uncover novel genomic signatures associated with non-linear processes that current theory has not yet suggested. Additionally, investigations on the robustness of neural networks to uncertain training data, and on new creative representation of population genetic data, will provide further opportunities for technological advancements in population genetics.
To conclude, we argue that the community should aim to make the field as inclusive as possible. If these conditions are met, deep learning will soon be established as part of the common toolkit among population geneticists globally.
This review is part of ongoing activities of the EvoGenomics.AI consortium.
Return to our blog